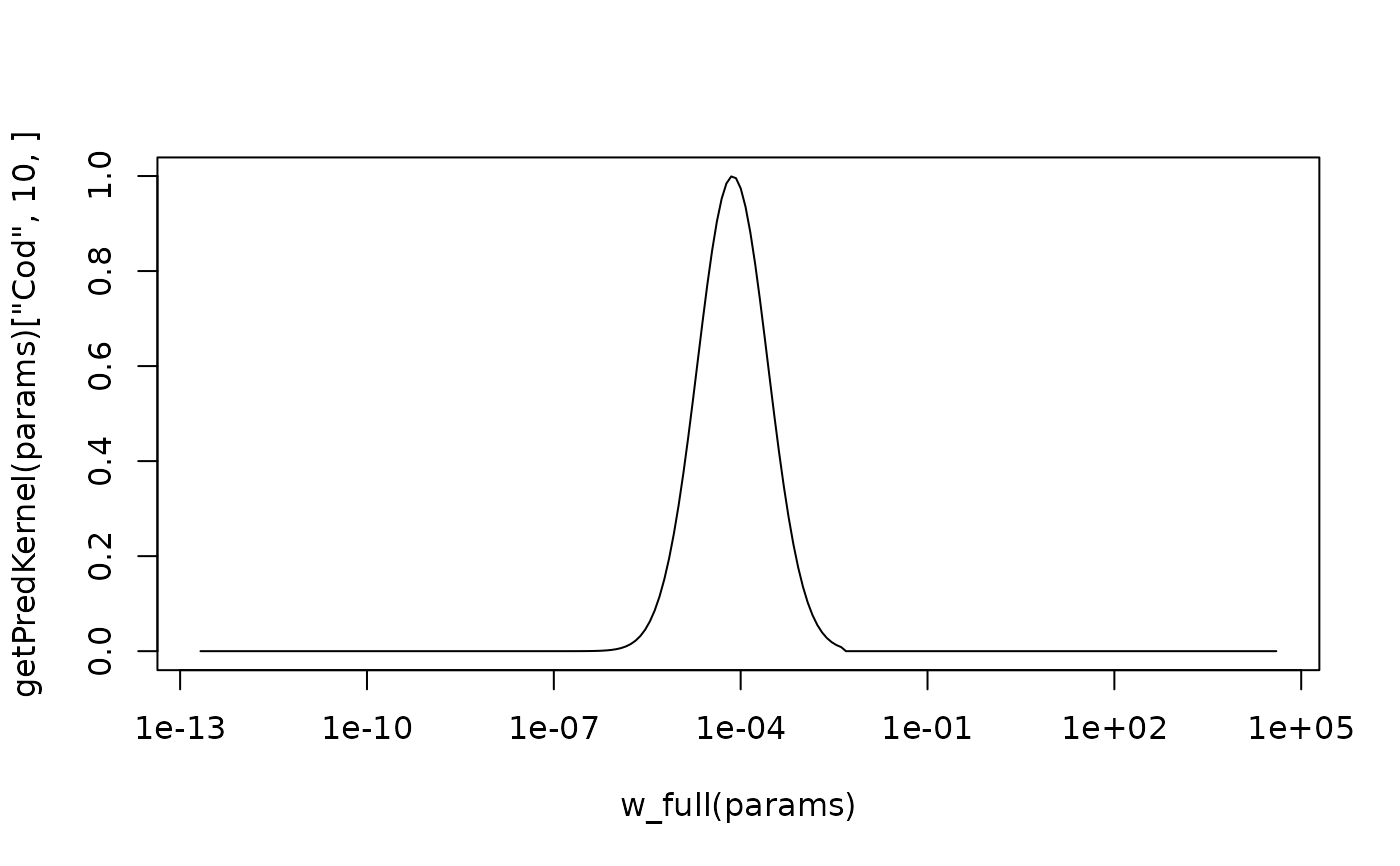
This predation kernel is a power-law, with sigmoidal cut-offs at large and small predator/prey mass ratios.
Arguments
- ppmr
A vector of predator/prey size ratios at which to evaluate the predation kernel.
- kernel_exp
The exponent of the power law
- kernel_l_l
The location of the left, rising sigmoid
- kernel_u_l
The shape of the left, rising sigmoid
- kernel_l_r
The location of the right, falling sigmoid
- kernel_u_r
The shape of the right, falling sigmoid
Value
A vector giving the value of the predation kernel at each of the
predator/prey mass ratios in the ppmr argument.
Details
The return value is calculated as
ppmr^kernel_exp /
(1 + (exp(kernel_l_l) / ppmr)^kernel_u_l) /
(1 + (ppmr / exp(kernel_l_r))^kernel_u_r)
The parameters need to be given as columns in the species parameter dataframe.
See also
Other predation kernel:
box_pred_kernel(),
lognormal_pred_kernel(),
truncated_lognormal_pred_kernel()
Examples
params <- NS_params
# Set all required paramters before changing kernel type
species_params(params)["Cod", "kernel_exp"] <- -0.8
species_params(params)["Cod", "kernel_l_l"] <- 4.6
species_params(params)["Cod", "kernel_u_l"] <- 3
species_params(params)["Cod", "kernel_l_r"] <- 12.5
species_params(params)["Cod", "kernel_u_r"] <- 4.3
species_params(params)["Cod", "kernel_type"] <- "power_law"
plot(w_full(params), getPredKernel(params)["Cod", 10, ], type="l", log="x")