Given a MizerParams object
params for which biomass observations are
available for at least some species via the biomass_observed column in the
species_params data frame, this function returns an updated MizerParams
object which is rescaled with scaleModel() so that the total biomass in
the model agrees with the total observed biomass.
Details
Biomass observations usually only include individuals above a certain size. This size should be specified in a biomass_cutoff column of the species parameter data frame. If this is missing, it is assumed that all sizes are included in the observed biomass, i.e., it includes larval biomass.
After using this function the total biomass in the model will match the
total biomass, summed over all species. However the biomasses of the
individual species will not match observations yet, with some species
having biomasses that are too high and others too low. So after this
function you may want to use matchBiomasses(). This is described in the
blog post at https://bit.ly/2YqXESV.
If you have observations of the yearly yield instead of biomasses, you can
use calibrateYield() instead of this function.
Examples
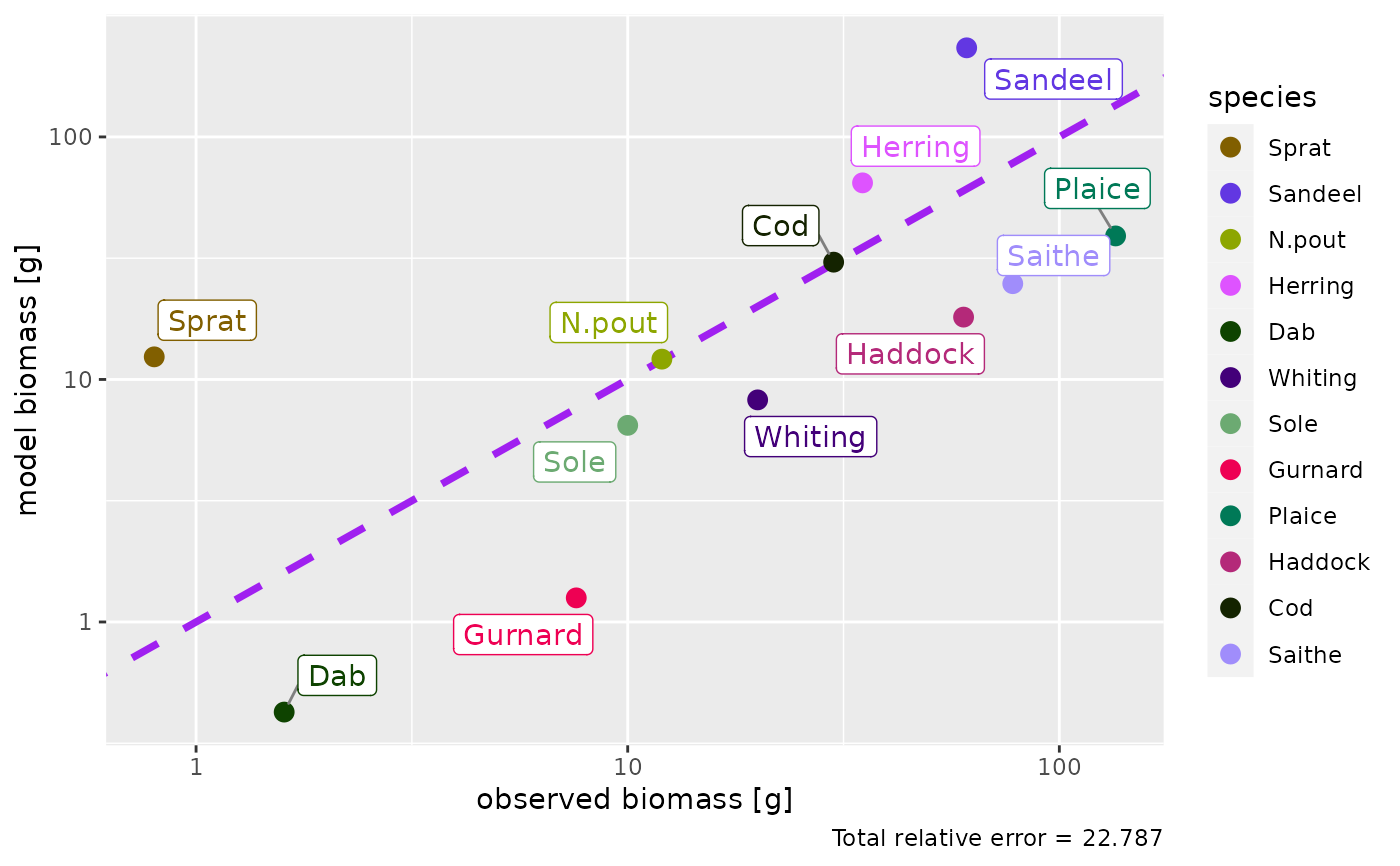
params <- NS_params
species_params(params)$biomass_observed <-
c(0.8, 61, 12, 35, 1.6, 20, 10, 7.6, 135, 60, 30, 78)
species_params(params)$biomass_cutoff <- 10
params2 <- calibrateBiomass(params)
plotBiomassObservedVsModel(params2)